Defining hierarchical protein interaction networks from spectral analysis of bacterial proteomes
If one were to buy a furniture set and accidentally lose the instruction manual, could we put the furniture back together? This is the question facing biologists all the time.
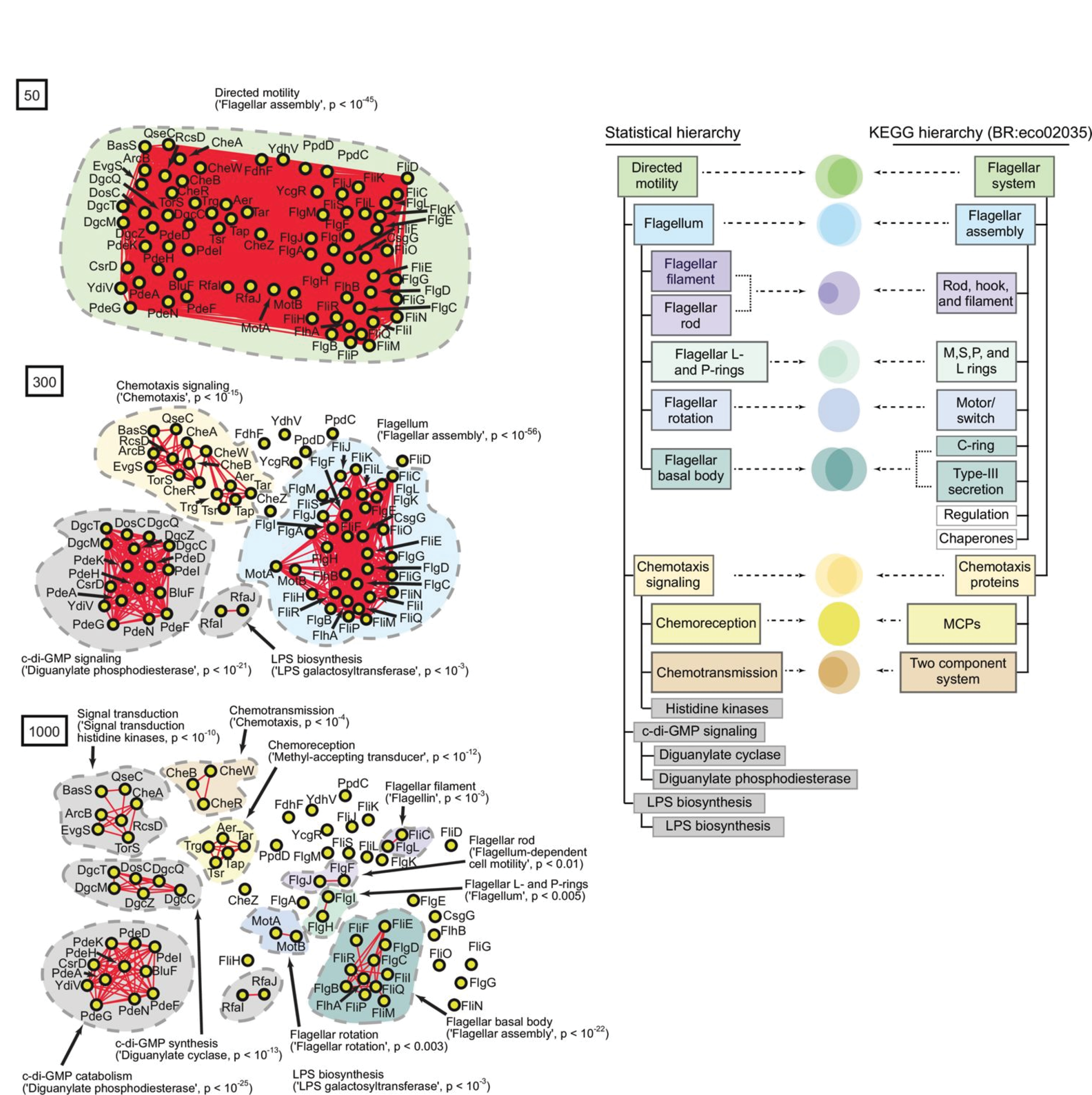
We inherit systems from nature that are built according to a process that is opaque to us and try to figure out their wiring diagram. If these systems have lots of parts, then this task becomes prohibitively difficult. Why? Imagine there are 100 components that make up the system. This means there are on the order of 2 100 or ~10 30 number of combinations that must be considered into order to understand the wiring diagram of the system. No number of robots could possibly get through this complexity. In this paper, we show that it is actually possible to use statistics infer the wiring diagram of complex biological systems. Key to this ability was the central insight that what has long been considered statistical noise in facts holds an incredible amount of signal that is more subtle and hierarchically organized. Thus, understanding emergence in biology—how individual parts integrate to create collective wholes—could now be possible simply from collecting data and performing statistical inference. This paper is the nucleating center of our laboratory, from which many projects have stemmed.